Examination of Genetic Background of Intrahepatic Cholangiocarcinoma by Bioinformatics Applications
Zeynep Kucukakcali1, Sami Akbulut21Department of Biostatistics and Medical Informatics, Inonu University Faculty of Medicine, Malatya, Türkiye2Department of Surgery and Liver Transplant Institute, Inonu University Faculty of Medicine, Malatya, Türkiye
Objectives: Intrahepatic cholangiocarcinoma (ICC), the second most common primary liver cancer, is associated with a poor prognosis with a very low survival rate. Therefore, a comprehensive understanding of the molecular pathways involved in the disease is of great importance for the development of targeted therapies and personalised treatment strategies. The aim of this study was to identify possible biomarkers associated with ICC by analysing gene expression in ICC tumor and non-tumour liver tissues.
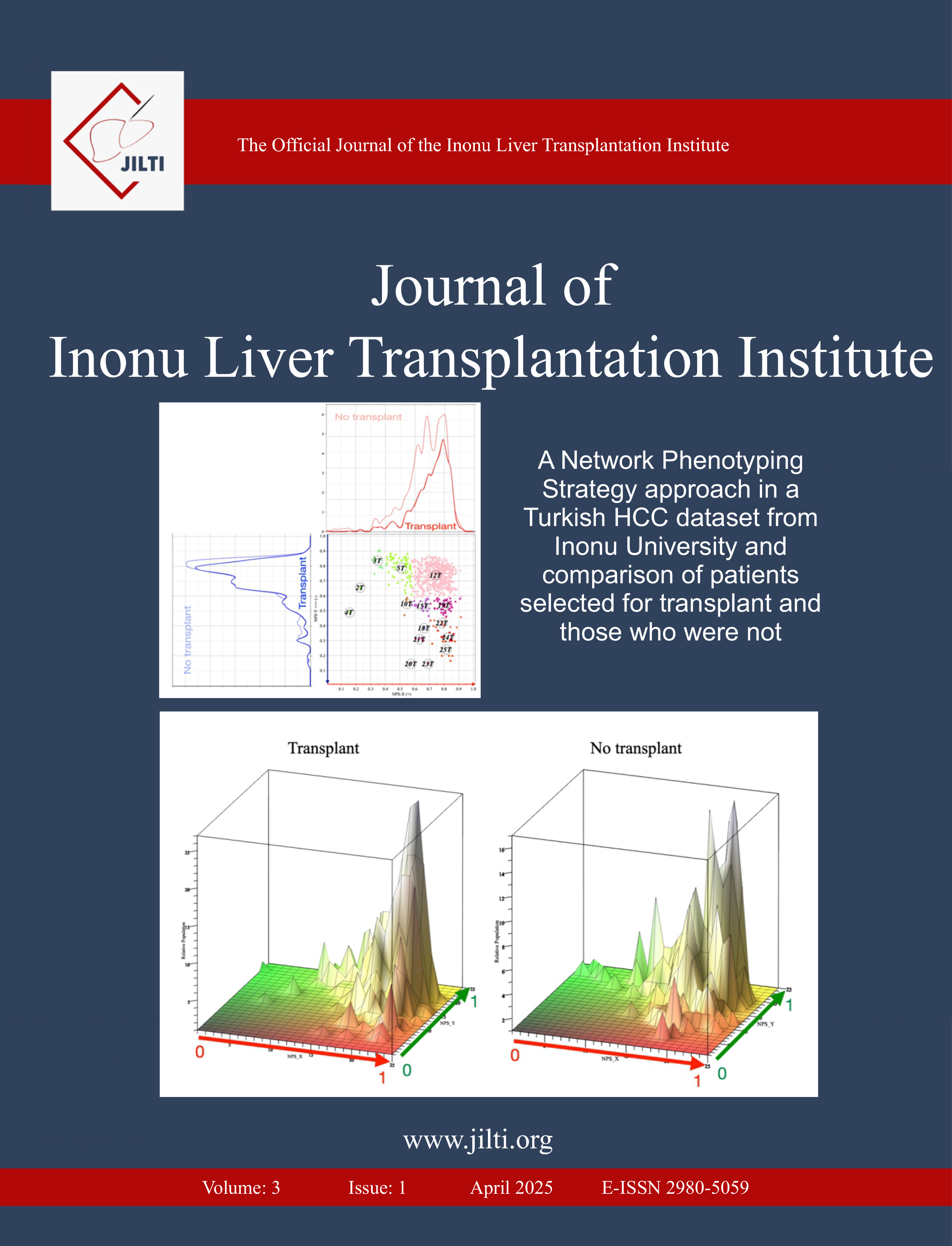
Methods: The dataset included in the study comprises gene expression data from ICC and non tumor liver tissue. The gene expression analysis of this data set was conducted using the capabilities provided by the limma package. The distribution of each tissue in the dataset is shown by the distribution graph. The UMAP graph represents the association of tissue types. The genes exhibiting different regulation are represented in the volcano plot.
Results: The UMAP analysis revealed a perfect separation of the tissues in the dataset into two distinct groups: ICC tumor tissues and non tumor liver tissues. The analysis showed that many genes differed in both groups under log2FC>1 p<0.05 and log2FC<-1 and p<0.05 conditions. The resultsshow that there are genes that are upregulated and down regulated in ICC tissues compared to non tumor liver tissues.
Conclusion: Genetic research has a pivotal role in enhancing investigate molecular pathways and treatment of ICC. Genes that have been identified can function as biomarkers, which can assist in the creation of medication therapies that are specifically targeted and enhance the quality of patient care and the efficiency of healthcare. As genetic research advances, the utilization of these biomarkers is anticipated to improve personalization.
Manuscript Language: English